Biography
Dr. Raviteja Vangara, Ph.D. is a Postdoctoral Researcher specializing in the advanced field of machine learning applications for scientific research. He currently holds a position at the esteemed Alexandrov lab within the Department of Cellular and Molecular Medicine at the University of California, San Diego (UCSD). Here, his research focuses on leveraging state-of-the-art machine learning methodologies to conduct insightful mutational signature analyses related to human cancer.
Before joining UCSD, Dr. Vangara contributed his extensive knowledge and innovative approach to research at the Theoretical Division of Los Alamos National Laboratory. His work at LANL covered a broad spectrum of scientific applications, predominantly utilizing unsupervised machine learning techniques such as graphical clustering methods, and non-negative matrix and tensor factorization techniques for advanced pattern recognition and latent feature extraction. During his tenure, Dr. Vangara played a crucial role in the Smart Tensors team, a 2021 R&D award winning collective responsible for releasing several groundbreaking open-source software tools optimized for high-performance computing scientific applications, focusing on scalable distributed computing methods.
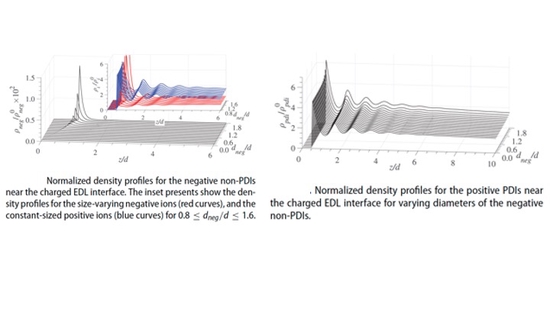
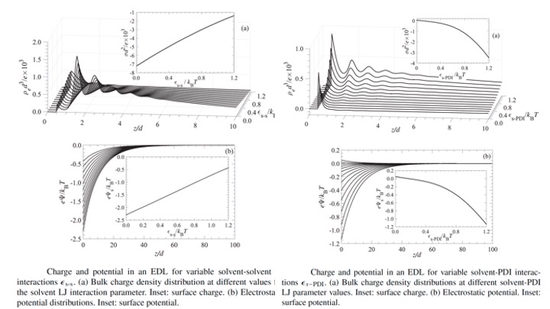
Dr. Vangara has been recognized for his academic excellence, earning his Ph.D. with distinction in 2019 for his significant work on Coulombic and non-Coulombic effects of Electric Double Layers. He obtained his M.S. in 2017 from the University of New Mexico’s Petsev lab, Department of Chemical and Biological Engineering, The University of New Mexico.
Download my resumé.
- Machine Learning
- Matrix and Tensor Factorization
- Cheminformatics and Bioinformatics
Ph.D. in Engineering (with Distinction), 2019
University of New Mexico
M.S. in Chemical Engineering, 2017
University of New Mexico
B.Tech. in Chemical Engineering, 2015
National Institute of Technology, Warangal
Recent News
May 2024: Geographic variation of mutagenic exposures in kidney cancer genomes paper published in Nature.
Dec 2023: SigProfilerAssignment paper published online on Oxford Bioinformatics.
Aug 2023: SigProfilerMatrixGenerator V2 paper published online on BMC Genomics.
Feb 2023: SigProfilerExtractor paper has been featured in Epidemiology and Genomics Research Program - EGRP’s Research Highlights for 2022.
Nov 2022: Article “Uncovering novel mutational signatures by de novo extraction with SigProfilerExtractor” published online.
Recent Publications
Featured Publications
Projects
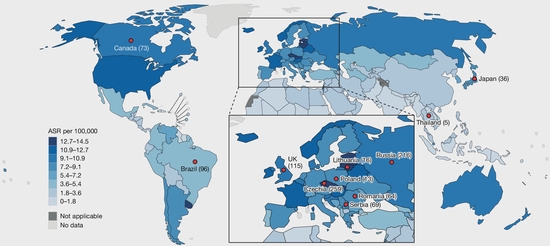
Geographic variation of mutagenic exposures in kidney cancer genomes
Geographic variation of mutagenic exposures in kidney cancer genomes
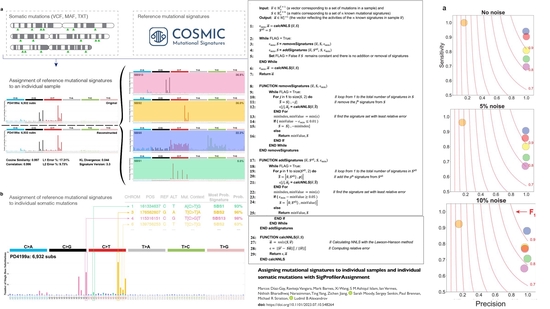
SigProfilerAssignment
Assigning mutational signatures to individual samples and individual somatic mutations with SigProfilerAssignment
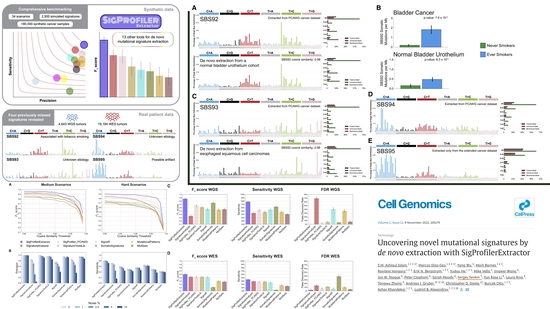
SigProfilerExtractor
Uncovering novel mutational signatures by de novo extraction with SigProfilerExtractor
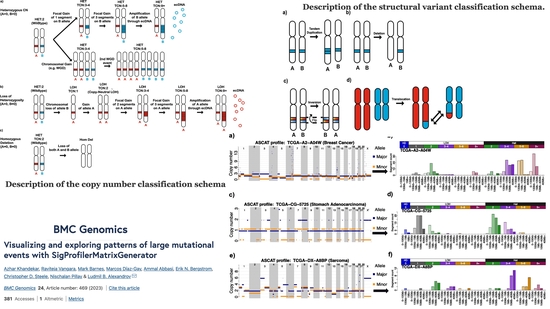
SigProfilerMatrixGenerator
Visualizing and exploring patterns of large mutational events with SigProfilerMatrixGenerator
The mutagenic forces shaping the genomic landscape of lung cancer in never smokers
The mutagenic forces shaping the genomic landscape of lung cancer in never smokers