SigProfilerMatrixGenerator
 SPMG2
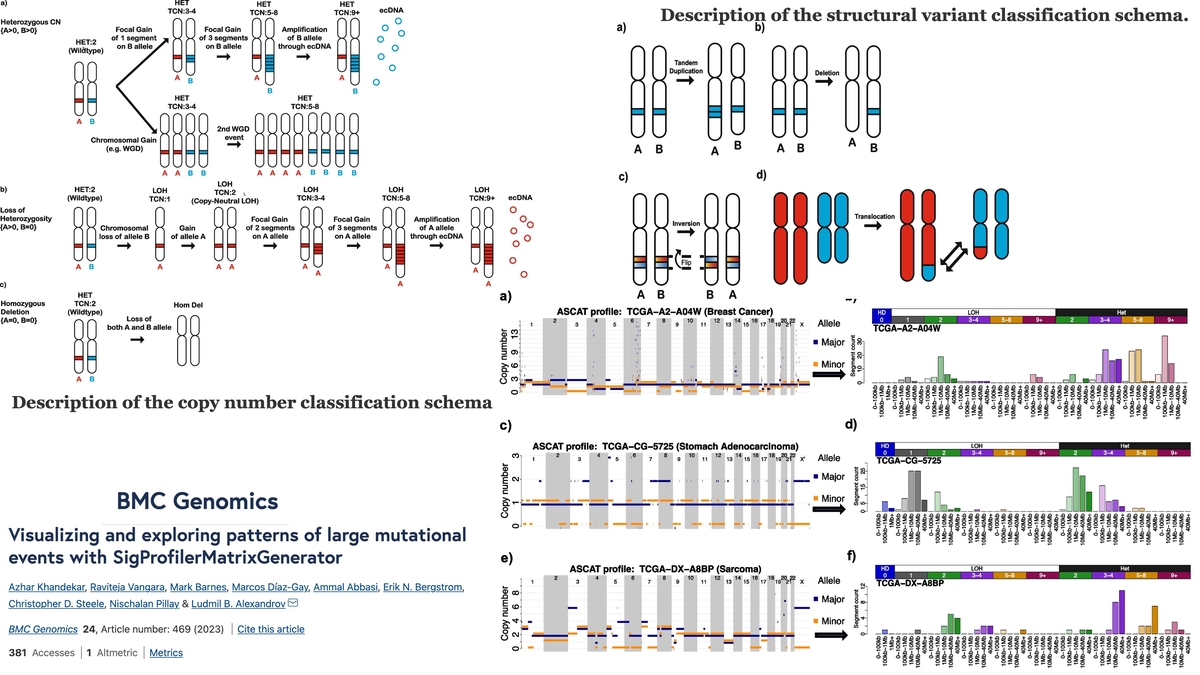
SPMG2All cancers harbor somatic mutations in their genomes. In principle, mutations affecting between one and fifty base pairs are generally classified as small mutational events. Conversely, large mutational events affect more than fifty base pairs, and, in most cases, they encompass copy-number and structural variants affecting many thousands of base pairs. Prior studies have demonstrated that examining patterns of somatic mutations can be leveraged to provide both biological and clinical insights, thus, resulting in an extensive repertoire of tools for evaluating small mutational events. Recently, classification schemas for examining large-scale mutational events have emerged and shown their utility across the spectrum of human cancers. However, there has been no computationally efficient bioinformatics tool that allows visualizing and exploring these large-scale mutational events.
Here, we present a new version of SigProfilerMatrixGenerator that now delivers integrated capabilities for examining large mutational events. The tool provides support for examining copy-number variants and structural variants under two previously developed classification schemas and it supports data from numerous algorithms and data modalities. SigProfilerMatrixGenerator is written in Python with an R wrapper package provided for users that prefer working in an R environment.
The new version of SigProfilerMatrixGenerator provides the first standardized bioinformatics tool for optimized exploration and visualization of two previously developed classification schemas for copy number and structural variants. The tool is freely available at https://github.com/AlexandrovLab/SigProfilerMatrixGenerator with an extensive documentation at https://osf.io/s93d5/wiki/home/.