Distributed NMFk
 Dnmfk
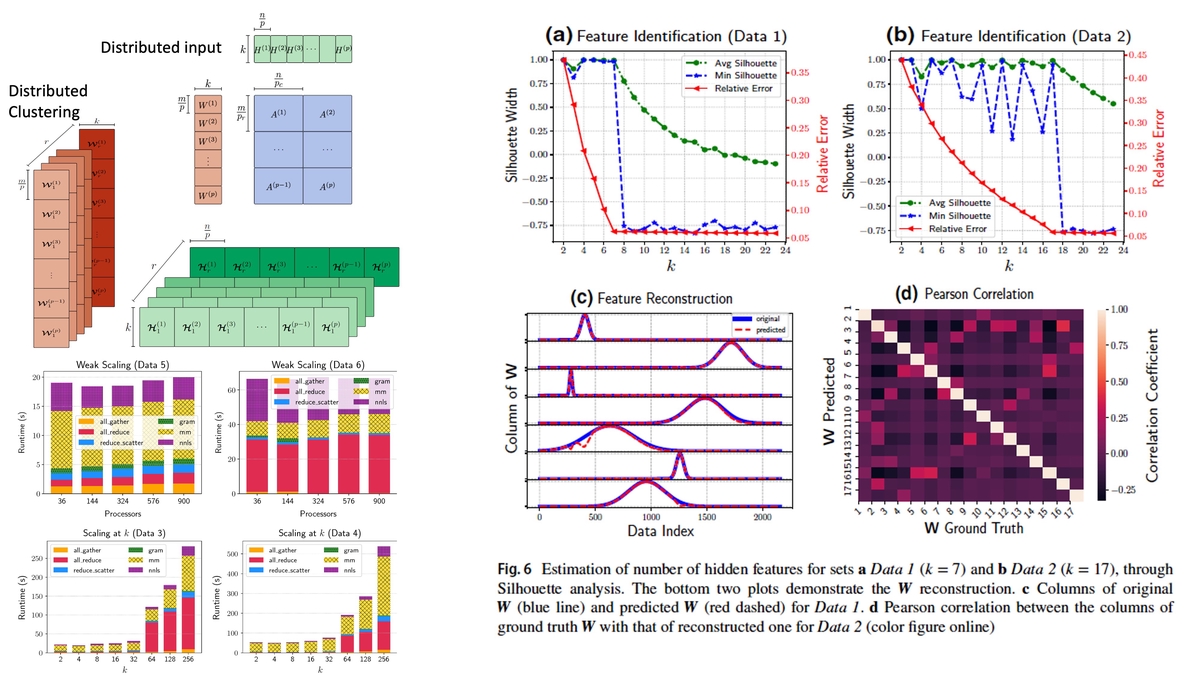
DnmfkThe holistic analysis and understanding of the latent (that is, not directly observable) variables and patterns buried in large datasets is crucial for data-driven science, decision making and emergency response. Such exploratory analyses require devising unsupervised learning methods for data mining and extraction of the latent features, and non-negative matrix factorization (NMF) is one of the prominent such methods. NMF is based on compute-intense non-convex constrained minimization, which, for large datasets requires fast and distributed algorithms. However, current parallel implementations of NMF fail to estimate the number of latent features. In practice, identifying these features is both difficult and significant for pattern recognition and latent feature analysis, especially for large dense matrices. In this paper, we introduce a distributed NMF algorithm coupled with distributed custom clustering followed by a stability analysis on dense data, which we call DnMFk, to determine the number of latent variables. The results on synthetic data and the classical Swimmer data set demonstrate the accuracy of model determination while scaling nearly linearly across multiple processors for large data. Further, we employ DnMFk to determine the number of hidden features from a terabyte matrix.